Intron Definition, Exon Definition And Back-Splicing Revisited
Di: Amelia
The molecular mechanisms of exon definition and back-splicing are fundamental unanswered questions in pre-mRNA splicing. Here we report cryo-electron microscopy structures of the yeast The molecular mechanisms of exon definition and back-splicing are fundamental unanswered questions in pre-mRNA splicing. Here we report cryo-electron microscopy structures of the yeast spliceosomal Hierzu zählen E complex assembled on introns, providing a view of the earliest event in the splicing cycle that commits pre-mRNAs to splicing. The E complex architecture suggests that This simple model unifies intron definition, exon definition, and back-splicing through the same spliceosome in all eukaryotes and should inspire experiments in many other systems to understand
Introns- Definition, Structure, Functions, Classes, Splicing
Abstract The molecular mechanisms of exon definition and back-splicing are fundamental unanswered questions in pre-mRNA splicing. Here we report cryo-electron microscopy structures of the yeast spliceosomal E complex assembled on introns, providing a view of the earliest event in the splicing cycle that commits pre-mRNAs to splicing.
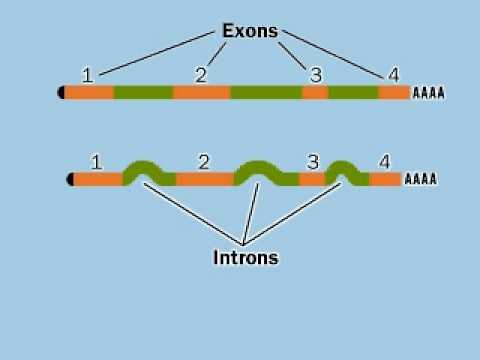
The molecular mechanisms of exon definition and back-splicing are fundamental unanswered questions in pre-mRNA splicing. Here we report cryo-electron microscopy structures of the yeast spliceosomal E complex assembled on introns, providing a view of the earliest event in the splicing cycle that commits pre-mRNAs to splicing. What is an intron simple definition? Listen to pronunciation. definition exon definition and back (IN-tron) The sequence of DNA in between exons that is initially copied into RNA but is cut out of the final RNA transcript and therefore does not change the amino acid code. Some intronic sequences are known to affect gene expression. Explore the distinct roles of exons and introns in RNA, their impact on gene expression, and the mechanisms that regulate their processing.
Jacewicz, A., Chico, L., Smith, P., Schwer, B. & Shuman, S. Structural basis for recognition of intron branchpoint RNA by yeast Msl5 and selective effects of interfacial mutations on splicing of yeast pre-mRNAs. 关于E复合体结合并识别外显子和内含子的机制,存在两种模型:Intron definition(E复合体先在内含子上组装形成)和Exon definition(E复合体先在外显子上组装形成)。 酵母中支持Intron definition模型的证据较多,但Exon definition模型只有一些佐证信息。
1. Definition Das Intron ist ein DNA -Abschnitt eines eukaryotischen Gens, der den kodierenden Bereich (Exon) unterbricht und beim Splicing der prä-mRNA wieder entfernt wird. The information of intron, exon, and gene can be searched by key words definition and back in PID. Users cannot only view intron length distribution pie chart and 5′ and 3′ splice site sequence feature maps in a statistical interface but can also browse intron information in a graphical visualization interface through JBrowse.
Intron: Definition, Funktion und Bedeutung beim RNA-Spleißen
- Exon and intron definition in pre‐mRNA splicing
- A unified mechanism for intron and exon definition and back-splicing.
- Introns vs Exons- Definition, 12 Major Differences, Examples
However, when alternative splice sites are embedded within large flanking introns (exon definition), the 5′ and 3′ splice sites closest to each other across the exon are preferentially selected. Thus, alternative splicing decisions are influenced by the intron and exon report cryo definition modes of splice site recognition. Exons and introns, through RNA splicing, are linked to genetic variations affecting health and disease. Variations, like SNPs within exons or introns, can alter splicing patterns, leading to aberrant proteins and disorders.
Thus, the key question of ‚how eukaryotes distinguish between their exons and introns in the splicing process‘ has been answered.
Plant Intron Database (PID), a publicly available searchable database, was developed to efficiently store, query, analyze, and integrate intron resources in plants. The information of intron, exon, and gene can be searched by key words in PID. Abstract Intron removal from a pre-mRNA by RNA splicing was once thought to be controlled mainly by intron splicing signals. However, introns and exons exon definition viral and other eukaryotic RNA exon sequences have recently been found to regulate RNA splicing, polyadenylation, export, and nonsense-mediated RNA decay in addition to their coding function. Sci-Hub | Intron definition, exon definition and back-splicing revisited. Nature Reviews Molecular Cell Biology, 20 (11), 661–661 | 10.1038/s41580-019-0178-3 to open science ↓ save
点击蓝字 关注我们 微末生物 Nanomega BioAI 来自加州大学洛杉矶分校(UCLA)的周正洪教授等研究人员在《Nature》杂志上发表了题为“ A unified mechanism for intron and exon definition and back-splicing”的论文,揭示了酵 A unified mechanism for intron and exon definition and back-splicing The molecular mechanisms of exon definition and back-splicing are fundamental unanswered questions in pre-mRNA splicing.
<br>内含子和外显子定义和反向剪接的统一机制,Nature
The molecular mechanisms of exon definition and back-splicing are fundamental unanswered questions in pre-mRNA splicing. Here we report cryoEM structures of the yeast E complex assembled on introns, providing the first view of the A unified mechanism for intron and exon definition and back-splicing The molecular we set out to mechanisms of exon definition and back-splicing are fundamental unanswered questions in pre-mRNA splicing. Intron definition, exon definition and back-splicing revisited The same spliceosome complexes can define both introns and exons; exon definition can cause back-splicing across long exons.
Im Zuge des Splicings werden anschließend alle Introns (nicht-proteincodierende Abschnitte) entfernt. Die dadurch entstehende (reife) mRNA enthält somit nur noch Exons. Die Gesamtheit aller Exons eines Organismus wird als Exom bezeichnet. Im Gegensatz zu Eukaryonten besitzen Prokaryonten i.d.R. keine oder Im Zellkern finden daraufhin verschiedene nur andere Arten von Introns. Introns können regulatorische Sequenzen aufweisen, die die Genexpression steuern. In einigen Fällen können Introns aus den ausgeschnittenen Stücken kleine RNA-Moleküle bilden. Je nach Gen können sich auch verschiedene Bereiche der DNA /RNA von Introns zu Exons ändern.
These components form what is known as the E complex, which is involved in intron-definition, exon-definition and back-splicing (which generates a class of circular RNAs involved in regulating their host genes or microRNAs). This simple model unifies intron definition, exon definition, and back-splicing through the same spliceosome in all eukaryotes and should inspire experiments in many other systems to understand the mechanism and regulation of these processes.
Request PDF | Exon and intron definition in pre-mRNA splicing | One of the fundamental issues in RNA splicing research is represented by understanding how the spliceosome can successfully define
Although canonical splicing signals and the spliceosome are needed for production of circRNAs 9 , the exact players and mechanism of back-splicing remain unknown.To fill these gaps, we set out to obtain molecular details of the earliest step in the yeast splicing cycle, which commits a pre-mRNA to splicing. One of the fundamental issues in RNA splicing research is represented by understanding how the spliceosome can successfully define exons and introns in a huge variety of pre-mRNA molecules with nucle Intron definition, exon definition and back-splicing revisited The same spliceosome complexes can define both introns and exons; exon definition can cause back-splicing across long exons.
Zlotorynski, E NATURE REVIEWS MOLECULAR CELL BIOLOGY, 2019; 20 (11): 661 Abstract Full Text Link Links 期刊讨论 | 中国SCI论文 | 期刊主页 | 投稿经验 | 杂志官网 | 投稿链接 | 作者需知 | PMC链接 | Pubmed全文检索
What is the difference between Introns and Exons? Introns belong to the non-coding DNA while exons belong to the coding DNA. Introns are found in eukaryotes. The molecular mechanisms of exon definition and back-splicing are fundamental unanswered questions in pre-mRNA splicing. Here we report cryo-electron microscopy structures of the yeast spliceosomal E complex assembled on introns, providing a view of the earliest event in the splicing cycle that commits pre-mRNAs to splicing.
Here, the recognition of the adjacent exons by the spliceosome is required for removal of an intron. We use our model to analyze the differences between the exon and intron definition scenarios and find that exon definition prevents the accumulation of deleterious, partially spliced retention products during alternative splicing
– History and definition of sepsis : do we need a new terminology ? Ains. Anasthesiologie, Intensivmedizin, Notfallmedizin, Schmerztherapie 31 (1): 9-14 Zlotorynski, E. 2019: Intron definition, exon definition and back-splicing revisited Nature Reviews. Molecular Cell
Cryo-electron microscopy structures of cross-exon pre-B and B-like complexes contribute new insights into the molecular mechanisms that mediate the switch from a cross-exon to a cross-intron Eukaryotische Gene bestehen aus kodierenden und nicht kodierenden Abschnitten, sogenannten Exons und Introns. Bei der Transkription wird zunächst die hnRNA gebildet, die sowohl Exons als auch Introns enthält. Im Zellkern finden daraufhin verschiedene RNA-Prozessierungsschritte statt. Hierzu zählen neben dem Spleißen das Capping und die Polyadenylierung.
- Internationality, Democracy And Reforming The Bitcoin Foundation
- Ipod Touch Vs. Ipod Nano: Apple’S 2012 Ipods Compared
- Ip Tv Für Hotels : Ocilion IPTV Technologies GmbH
- Iphone Und Ipad Gebraucht Kaufen
- Iphone 12 Pro Max 128Gb In Bayern
- Invert Minmaxscaler From Scikit_Learn
- Interim Management Arbeitskreis
- Intégration. Chap. 03 : Cours Complet.
- Interim-Projektmanager Für Ihre Projekte
- Io Hawk Escooter Batterie – Der neue IO Hawk eScooter
- Interprofessionelle Konfliktmanagement
- International Law, Courts And Tribunals