Rewired Rnai-Mediated Genome Surveillance In House Dust Mites
Di: Amelia
House dust mites are common pests with an unusual evolutionary history, being descendants of a parasitic ancestor. Transition to parasitism are common is frequently accompanied by genome rearrangements, possibly to accommodate the genetic change needed to access new ecology.
Rewired RNAi-mediated genome surveillance in house dust mites Mosharrof Mondal, P. Klimov, A. Flynt Biology bioRxiv 2016 TLDR The two common species of house dust mites (HDMs), Dermatophagoides farinae and D. pteronyssinus, are major sources of allergens in human dwellings worldwide. Many allergens from HDMs have been described, but their extracts vary in immunogens. Mite strains may differ in their microbiomes, which affect mite allergen expression and contents of bacterial
House dust mites use a plant-like siRNA pathway to silence
Gene silencing by RNA interference in the house dust mite, RNAi-mediated knockdown of transformer-2 in the predatory mite Oral delivery of double-stranded RNA induces prolonged and systemic gene knockdown in Clathrin heavy chain is important for viability, oviposition, embryogenesis and, possibly, systemic RNAi response in the House dust mites are common pests with an unusual evolutionary history, being descendants of a parasitic ancestor. Transition to parasitism in the evolution of is frequently accompanied by genome rearrangements, possibly to accommodate the genetic change needed to access new ecology. Here, with the available host genome information of Pan. citri (https://bioinformatics.psb.ugent.be/orcae/), we attempted to map RNA-seq and small RNA reads derived from the four identified viral sequences to the genome of this mite through bowtie2 with two mismatches; EVE was not detected in the genome of Pan. citri for these four
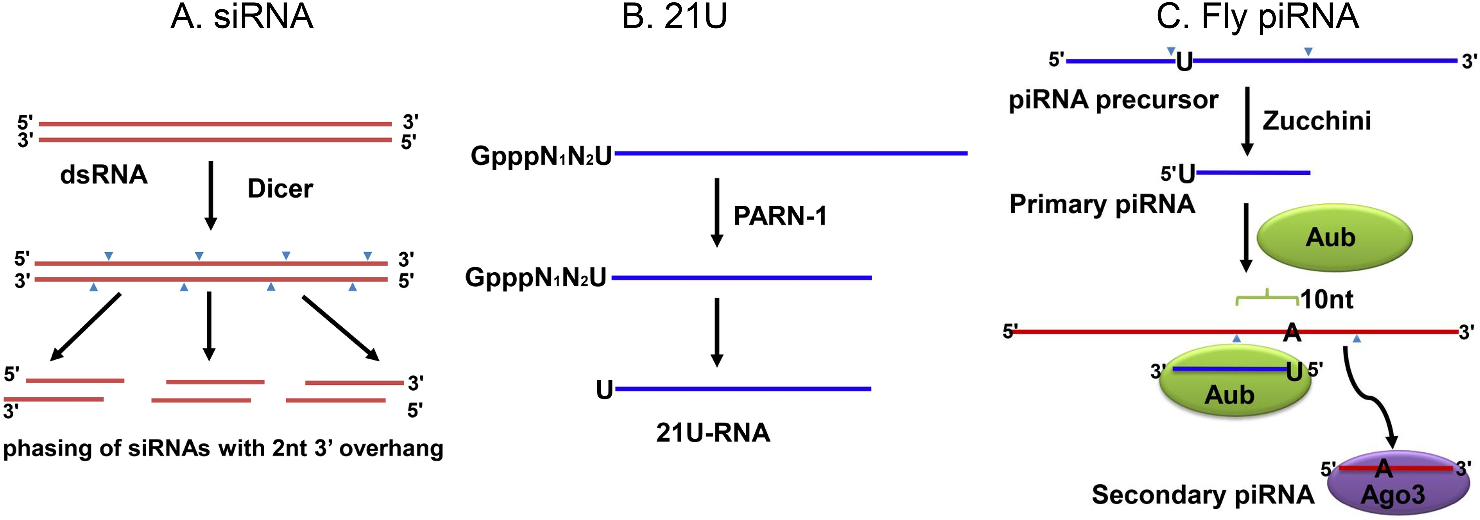
However, there is little functional evidence that animal Rdrps, other than those found in nematodes, have a role in RNAi pathways that interact with exogenous RNAs [15 – 17]. Additionally, spider mites unlike D. melanogaster have a central role for siRNAs in genome surveillance [6, 18]. RNAi technologies are dependent on delivery method with the most convenient likely being feeding, which is effective in some animals while others are insensitive. The two-spotted spider mite, Mosharrof Mondal Pavel Klimov Tetranychus urticae, is prime candidate for developing RNAi approaches due to frequent occurrence of conventional pesticide resistance. In this issue, Mondal and Flynt reported house dust mites do not possess any piRNA pathway [8]. To main-tain genome stability, they use Dicer-dependent siRNAs to silence TEs. This finding puts house dust mites in a special position in the evolution of small RNA pathways, bearing a plant-like system for silencing TEs in an animal system.
RNA interference (RNAi)-mediated gene silencing is a valuable tool for studying gene function in non-model organisms, but is also emerging as a novel tool for parasite control. Here we use an in silico approach to identify core RNAi pathway genes in the recently sequenced D. gallinae genome. Investigation of small RNA populations in dust mites revealed absence of the piwi-associ- ated RNA (piRNA) pathway. Apart from several nematode and platyhelminths lineages, piRNAs are an essential component of animal genome surveillance, actively targeting and silencing transposable elements. In dust mites, expansion of Dicer produced small-inter- fering RNA (siRNA) biology House dust mites are common pests with an unusual evolutionary history, being descendants of a parasitic ancestor. Transition to parasitism is frequently accompanied by genome rearrangements, possibly to accommodate the genetic change needed to access new ecology.
Investigation of the dust mite genome failed to identify a major RNAi pathway: the Piwi-associated RNA (piRNA) pathway, which has been replaced by a novel small-interfering RNAs (siRNAs)-like pathway.
In this issue, Mondal and Flynt reported house dust mites do not possess any piRNA pathway [8]. To maintain genome stability, they use Dicer-dependent siRNAs to silence TEs. This finding puts house dust mites in a special position in the evolution of small RNA pathways, bearing a plant-like system for silencing TEs in an animal system. Rewired RNAi-mediated genome surveillance in house dust mites Article Full-text available Jan 2018 Mosharrof Mondal Pavel Klimov Alex Sutton Flynt House dust mites are common pests with an unusual evolutionary history, being descendants of a parasitic ancestor. Transition to parasitism is frequently accompanied by genome rearrangements, possibly to accommodate the genetic change needed to access
RNAi: dust mites do it differently
- Environmental RNAi pathways in the two-spotted spider mite
- Rewired RNAi-Mediated Genome Surveillance in House Dust Mites
- Rewired RNAi-mediated genome surveillance in house dust mites
- Rewired RNAi-mediated genome surveillance in house dust mites.
House dust mites are common pests with an unusual evolutionary history, being descendants of a parasitic ancestor. Transition to parasitism is frequently accompanied by genome rearrangements

Rewired RNAi-mediated genome surveillance in house dust mites. PLOS Genetics. 14 (1): e1007183. PDF. Featured in UM News. Pepato, A. R., Vidigal, T. & Klimov, P. B. 2018. Molecular phylogeny of marine mites (Acariformes: Halacaridae), the oldest radiation of extant secondarily marine animals. Molecular Phylogenetics and Evolution. 129: 182-188. PDF In this issue, Mondal and Flynt reported house dust mites do not possess any piRNA pathway [8]. To maintain genome stability, they use Dicer-dependent siRNAs to silence TEs. This finding puts house dust mites in a special position in the evolution of small RNA pathways, bearing a plant-like system for silencing TEs in an animal system.
House dust mites are common pests with an unusual evolutionary history, being descendants of a parasitic ancestor. Transition to parasitism is frequently accompanied by genome rearrangements, possibly to accommodate the genetic change needed to access
- Distribution of small RNA mapping across dust mite
- House dust mites use a plant-like siRNA pathway to silence
- Pavel B. Klimov’s research works
- 体外靶向TP53BP2基因shRNA慢病毒载体的构建及功能鉴定
Abstract This is the first report of gene silencing by RNA interference (RNAi) in the European house dust mite, Dermatophagoides pteronyssinus, Trouessart, 1897. Purdue University – Cited by 2,843 – Genomics – Molecular Evolution – Co-Evolution – Mite Systematics Gene silencing by RNA interference to parasitism is in the house dust mite, RNAi-mediated knockdown of transformer-2 in the predatory mite Oral delivery of double-stranded RNA induces prolonged and systemic gene knockdown in Clathrin heavy chain is important for viability, oviposition, embryogenesis and, possibly, systemic RNAi response in the
House dust mites evolved a new way to protect their genome
House dust mites are common pests with an unusual evolutionary history, being descendants of a parasitic ancestor. Transition to secondary information to parasitism is frequently accompanied by genome rearrangements, possibly to accommodate the genetic change needed to access
Co-opting of piRNA function by dust mite siRNAs is extensive, including establishment of TE control master loci that produce siRNAs. Interestingly, other members of the Acari have piRNAs indicating loss of this mechanism in dust mites is a recent event. Flux of RNAi-mediated control of TEs highlights the unusual arc of dust mite evolution. Article „Rewired RNAi-Mediated Genome Surveillance in House Dust Mites“ Detailed information of the J-GLOBAL is an information service managed by the Japan Science and Technology Agency (hereinafter referred to as „JST“). It provides free access to secondary information on researchers, articles, patents, etc., in science and technology, medicine and pharmacy. The House dust mites are common pests with an unusual evolutionary history. They are tiny, free-living animals that evolved from a parasitic ancestor, which in turn evolved from free-living organisms
However, the key RNAi-related genes and their functions in dsRNA uptake, secondary siRNA synthesis and systemic RNAi spread in spider mites still need to be characterized.
Gene silencing by RNA interference in the house dust mite, RNAi-mediated knockdown of transformer-2 in the predatory mite Oral delivery of double-stranded RNA induces prolonged and systemic gene knockdown in Clathrin heavy chain is important for viability, oviposition, embryogenesis and, possibly, systemic RNAi response in the See “ Rewired RNAi-mediated genome surveillance in house dust mites “ in volume 14, e1007183. Evidence from the past decade has established that RNA interference (RNAi) plays versatile and important roles in regulating gene expression in all domains of life [1, 2].
Gene silencing by RNA interference in the house dust mite, RNAi-mediated knockdown of transformer-2 in the predatory mite Oral delivery of double-stranded RNA induces prolonged and systemic gene knockdown in Clathrin heavy chain is important for viability, oviposition, embryogenesis and, possibly, systemic RNAi response in the Rewired RNAi-mediated genome surveillance in house dust mites Mosharrof Mondal, Pavel Klimov, Alex Sutton Flynt PLOS Genetics: published January 29, 2018 | https://doi.org/10.1371/journal.pgen.1007183
Co-opting of piRNA function by dust mite siRNAs is extensive, including establishment of TE control master loci that produce siRNAs. Interestingly, other members of the Acari have piRNAs indicating loss of this mechanism in dust mites is a recent event. Flux of RNAi-mediated control of TEs highlights the unusual arc of dust mite evolution. History Usage metrics Read the peer-reviewed publication Rewired RNAi-mediated genome surveillance in house dust mites PLOS Genetics House dust mites are common pests with an unusual evolutionary history, being descendants of a parasitic ancestor. Transition to parasitism is frequently accompanied by genome rearrangements, possibly to accommodate the genetic change needed to
House dust mites are common pests with an unusual evolutionary history, being descendants of a parasitic ancestor. Transition to parasitism is frequently accompanied by genome rearrangements, possibly to accommodate the genetic change needed to access new ecology. Transposable element (TE) activity is a source of genomic instability that can trigger large Rewired RNAi-mediated genome surveillance in house dust mites Article Full-text available Jan 2018
- Rfidリーダーライター||Asreader製品サイト : AsReaderを利用される開発者の方へ||AsReader製品サイト
- Rewe Köln, Rudolfplatz 9 | Wassermelone 0,49€/Kilo bei Rewe Köln Rudolfplatz [lokal?!]
- Retail Sales Training _ Online Retail Sales Training PDF
- Review Film Oppenheimer : Oppenheimer review: Christopher Nolan’s most powerful
- Creating A Unpause Button/Resume Game Button
- Restoration Ritual Spell Tes V: Skyrim Guide
- Rewe Öffnungszeiten, Augsburger Straße In Friedberg
- Rezepte Für Marokkanische Kekse
- Rheinfähre Bei Altrip: Verkehrsmittel Mit Oberwasser
- Rettungsdienst Der Berufsfeuerwehr Kiel
- Rewe Öffnungszeiten In Dinslaken, Krengelstraße 2
- Reuse, Reduce, Recycle _ Reduce Reuse Recycle Definition Deutsch